About PhenomeCentral
Contents
- Contents
- Overview
- Who is contributing data to PhenomeCentral?
- Setting up an account
- Using PhenomeCentral
- PhenomeCentral exists thanks to
- Citing PhenomeCentral
- More questions?
Overview
PhenomeCentral is a repository for secure data sharing targeted at clinicians and scientists working in the rare disorder community. We encourage global scientific collaboration while respecting the privacy of patients profiled in this centralized database.
- PhenomeCentral enables the discovery of multiple individuals affected by the same unnamed disorder
- The same undiagnosed disorder may be present in only a handful of individuals seen at different hospitals from different corners of the world. The PhenomeCentral collaboration model allows clinicians and scientists to learn about the existence of cases similar to theirs, which may help to improve understanding of disorder manifestations and confirm underlying causes.
- The matching algorithms are sensitive to atypical phenotypic manifestations of disorders
- The semantic model, which is the basis of our matching process, helps reveal subtle similarities between disorder manifestations and may provide justification for reconsidering diagnoses that were initially discarded due to atypical phenotypes.
Who is contributing data to PhenomeCentral?
Several major rare disease research programs, including the Canadian CARE for RARE Project, the RD-Connect Project and the NIH Undiagnosed Diseases Program, are contributing interesting cases to PhenomeCentral. Together, we are building an extensive rare disorder knowledge base, and are establishing collaborations in the search for the underlying genetic causes of unsolved cases. PhenomeCentral currently has 6416 members, who have contributed hundreds of cases that are available for phenotype matching. Additionally, all registered users have view-only access to cases that have been made public by other users.
Setting up an account
Please use the SIGN UP button on the homepage to request a PhenomeCentral account. You will be asked to provide your name, affiliation, an institutional email address, and confirmation that you intend to contribute real de-identified data to PhenomeCentral. The identity of all prospective users requesting accounts is verified before granting them access to PhenomeCentral. It may take a few days for your request to be accepted. This process is a part of our efforts to ensure that the PhenomeCentral database is populated only with real, high-quality profiles of patients with rare disorders.
Using PhenomeCentral
Registered PhenomeCentral users can
- view available public data,
- contribute new cases,
- discover patients demonstrating similar phenotypes, which may help users gain more insight into or even solve undiagnosed cases,
- establish collaborations,
- share scientific information with other contributors.
Browsing the available data
Once you are logged in, the PhenomeCentral homepage provides quick access to all information of interest:
Left column: your data
- The MY DATA box lists the patient records owned by the current user. Please read below for more information about data ownership.
- The DATA SHARED WITH ME box lists the patient records for which the current user is a collaborator either because he or she is part of the group which owns the record, or because the owner explicitly listed them as a collaborator on that record. Please read below for more information about specifying collaborators.
Right column: public data
- The PUBLIC DATA box lists the patient records that are available for viewing to all registered users.
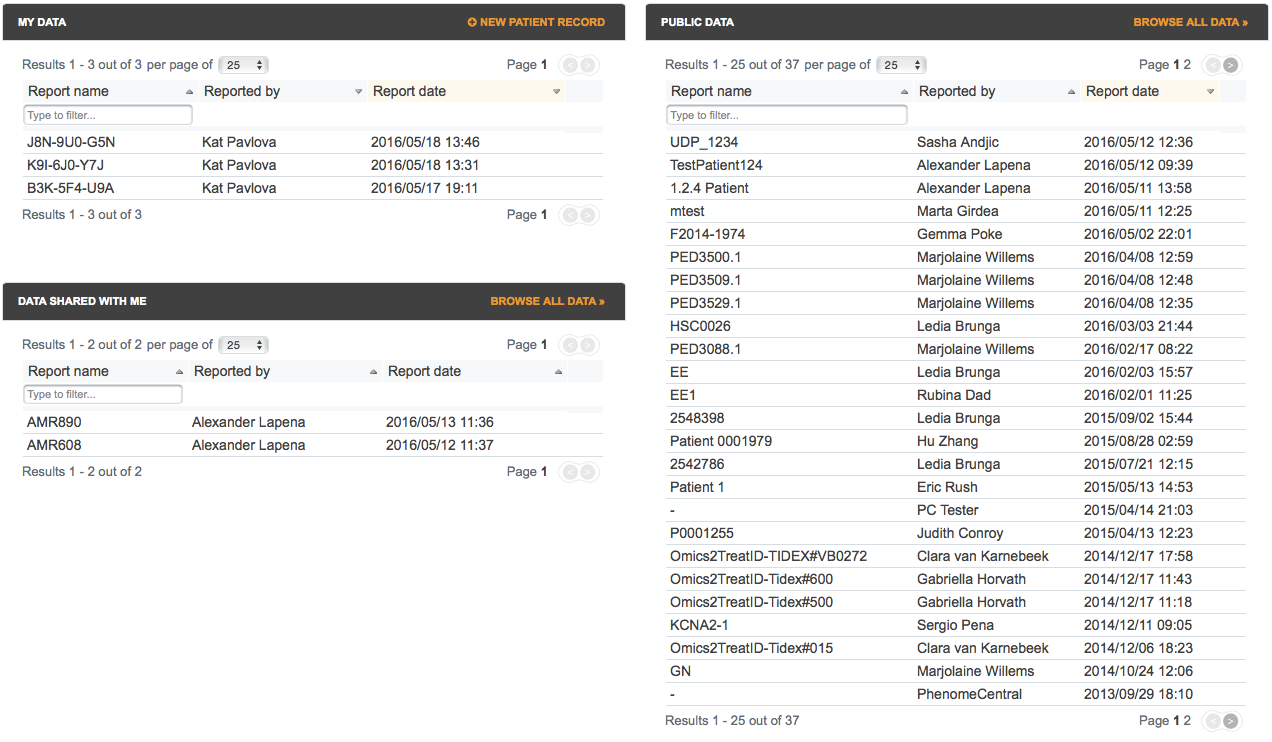
The homepage is immediately accessible from any other page by clicking on the PhenomeCentral logo or on the home icon ( ) displayed at the bottom of the page header area.

Data entry
PhenomeCentral relies on the PhenoTips software for data entry and storage. PhenoTips enables data capture and storage using the standardized vocabulary defined by the Human Phenotype Ontology (HPO). PhenomeCentral relies on the semantic phenotype relations defined by the ontology to identify the most relevant patient profile matches and to help users discover clusters of patients that may suffer from the same disorder.
Phenotypes can be selected from a predefined list of common manifestations, or by searching the entire ontology using the Quick Phenotype Search field for the relevant manifestations that users have observed in their patients. A phenotype can be marked as observed (Y), not observed and relevant (N), or not observed and/or not investigated, i.e. irrelevant (NA). It is not necessary to specify that a phenotype listed in the form is NOT present, unless the information about its absence is considered particularly relevant. Each phenotype can be annotated with information such as age of onset and pace of progression, which will help narrow down the possible diagnoses and find the most relevant phenotype matches in the database. It is important to know that while free-form text can be entered, these entries are not searchable or matchable within the registry. Therefore, users are encouraged to provide detailed phenotypic descriptions using ontology terms, as more precision will lead to a more accurate knowledge base for the community, as well as to better case match results for the contributor.
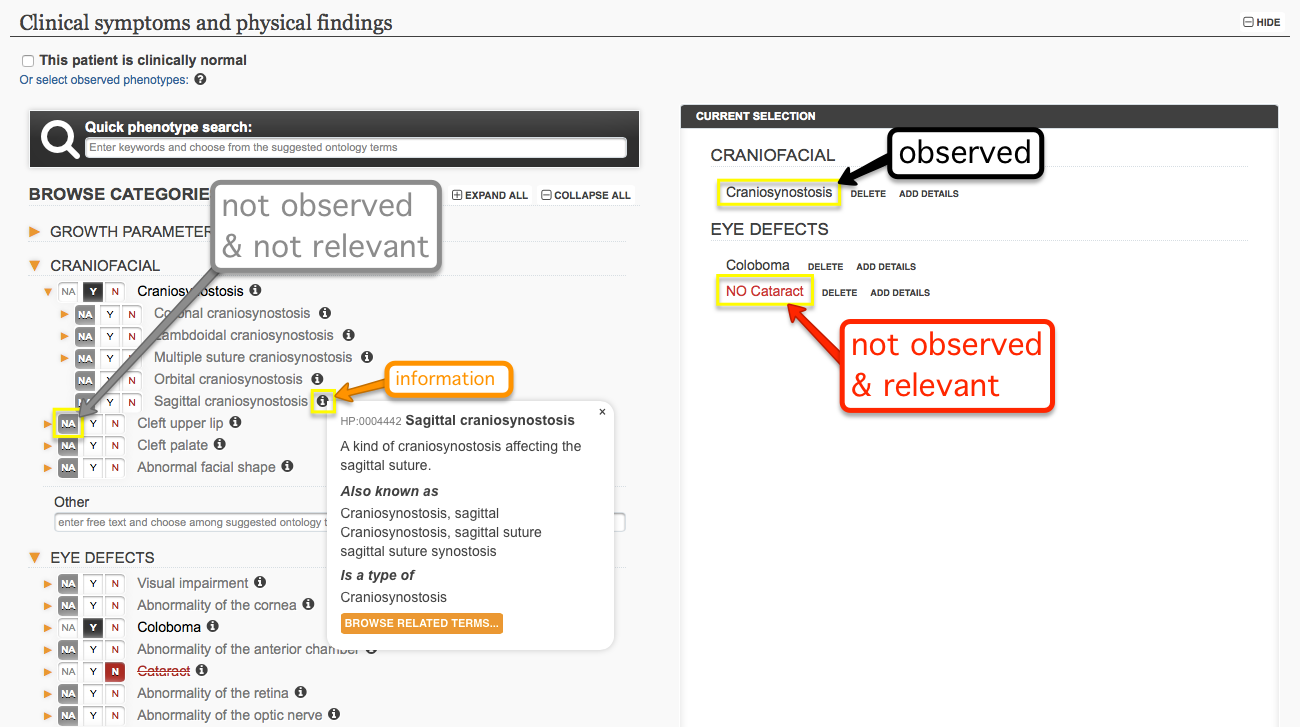
Entries owned by a contributor can be accessed at any time via the "MY DATA" box displayed on the homepage. Users can also modify or delete the patient profiles via the patient report menu available on the right side of the page.
Permissions and data sharing
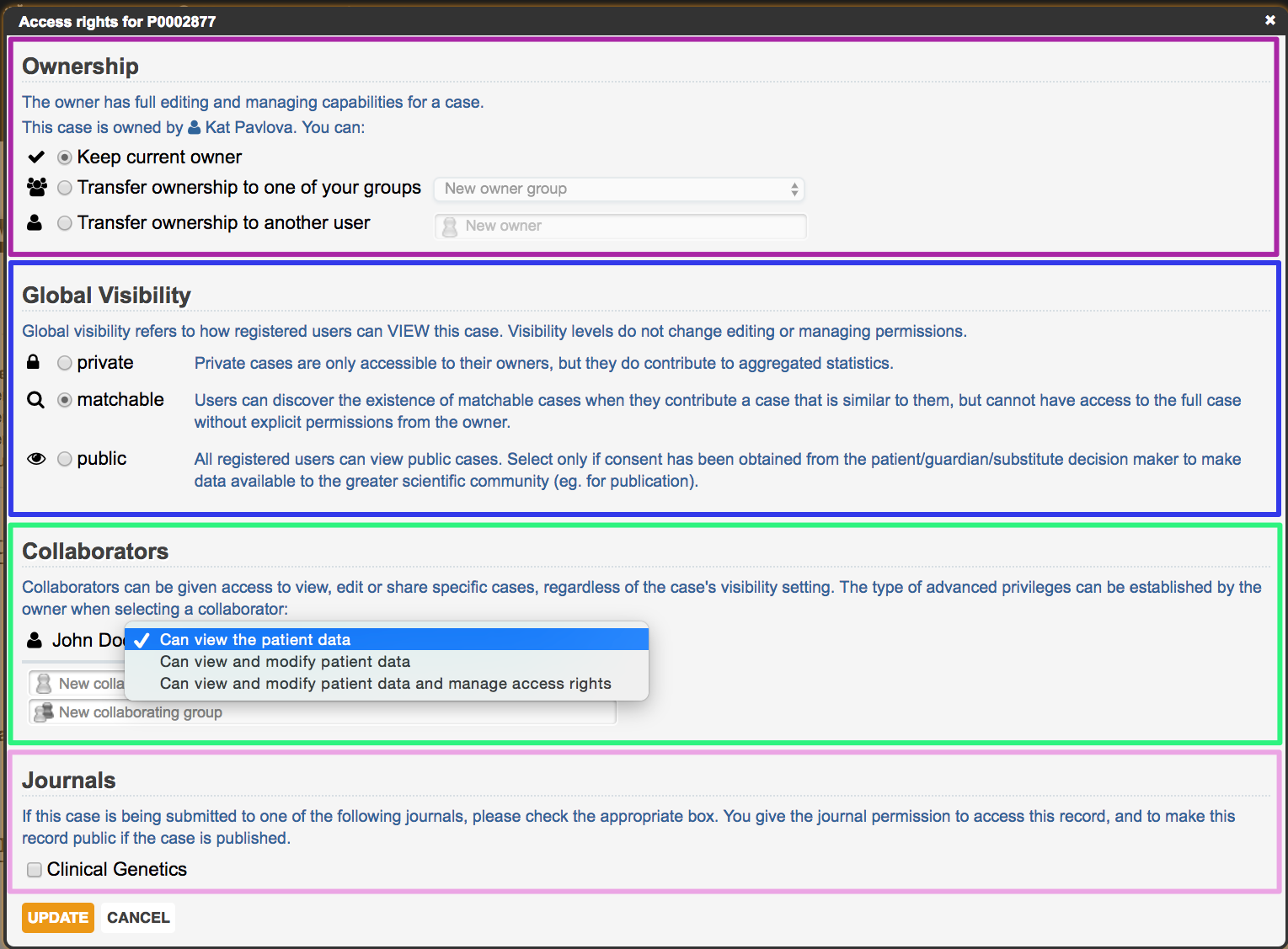
Patient record ownership, visibility, and collaboration can all be managed within the Modify Permissions setting, which is found in the upper-right area of the patient record. As well, if you are using PhenomeCentral for the purpose of submitting a case to a journal, you can share the record with them through this setting.
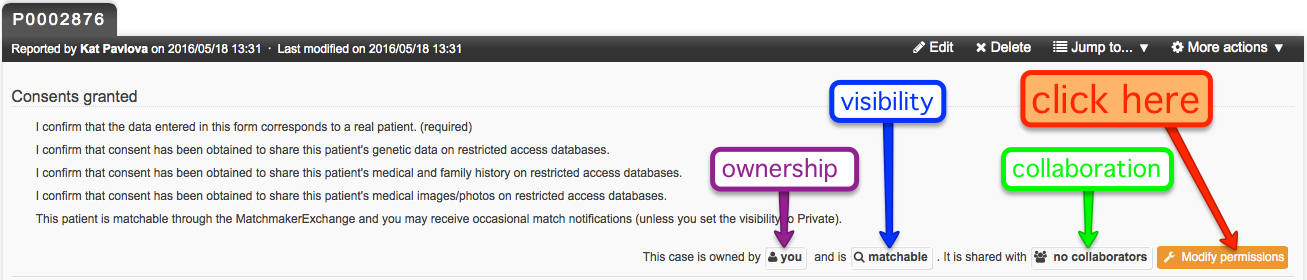
Case ownership
By default, you are the owner of any data that you have entered in the repository. Owners of a patient record can edit the record, choose to share the record with collaborators, and manage the record. Under certain circumstances, you may wish to transfer ownership of a patient record to a larger consortium if you and your case become part of a wider study, or to another PhenomeCentral contributor if for any reason they become responsible for following that specific patient.
When viewing or editing a patient record, the owner is displayed at the top of the page. Ownership can be modified by the owner, i.e. transferred to one of the owner's groups or to another user.
Establishing visibility
PhenomeCentral supports three major levels of data visibility:
- private: hidden from everyone except the owner and their explicit collaborators, but patient data is allowed to contribute to aggregated statistics
- matchable: the record cannot be directly viewed or searched by registered users, but is available to the automated phenotype matching algorithm. This allows for registered users to be aware of the case if it is a match for other similar cases. See Discovering similar cases
- public: viewable and searchable by all registered users
By default, cases are set to be matchable.
When viewing or editing a patient record, the visibility is displayed at the top of the page. Visibility can be modified both by the record owner and by those collaborators that have permissions to manage access rights.
Adding collaborators to your cases
Adding collaborators allows you to give a trusted colleague access to one of your non-public cases, as well as optional editing capabilities to any of your cases.
When viewing or editing a patient record, the number of collaborators is displayed at the top of the page. Collaborators can be awarded different levels of access to the record, ensuring the collaborator's ability to either:
- view the full patient profile
- view and modify the patient data
- view and modify the patient data, as well as manage access rights (referring to the ability to fully modify collaborator, visibility, and journal settings)
Submitting a patient record to a journal
Some journals require cases to be submitted to a restricted-access database as a condition of publication. If you are submitting a patient record to a journal that has partnered with PhenomeCentral, you can easily grant the journal access to this record by selecting the journal from the Journals section of the permissions popup.
The Modify permissions popup, with the default settings selected for each of the above four categories:
Discovering similar cases in PhenomeCentral
Owners of matchable or public records can see the existing matchable and public cases that are most similar to theirs. PhenomeCentral uses an algorithm that assesses the similarity of accessible cases based on the provided phenotypes (i.e. the selected Human Phenotype Ontology (HPO) terms). The algorithm finds cases with similar terms based on the Human Phenotype Ontology (HPO), prioritizing matches with more similar terms and more informative terms (i.e. rare phenotypes as assessed by numerous sources). For example, cases that share the term "oculomotor apraxia" would be a better match than cases that share the term "apraxia", while the algorithm will recognize that "intellectual disability" and "global developmental delay" are terms that share some phenotypic similarity, even though they are not exact matches.
After creating a patient record, contributors can see the most similar cases found in the database displayed at the bottom of the page. The list is re-generated each time the user accesses the patient record and may change as data is modified for that record or for other records in the database.
Private and hidden cases do not participate in any way in the similarity matching process. If your patient record is private or hidden, you will not be able to see similar cases, and your case will not be included in the list of matching similar cases for any other users. If you wish to benefit from the system's similarity feature, please consider giving your records at least the matchable visibility level.
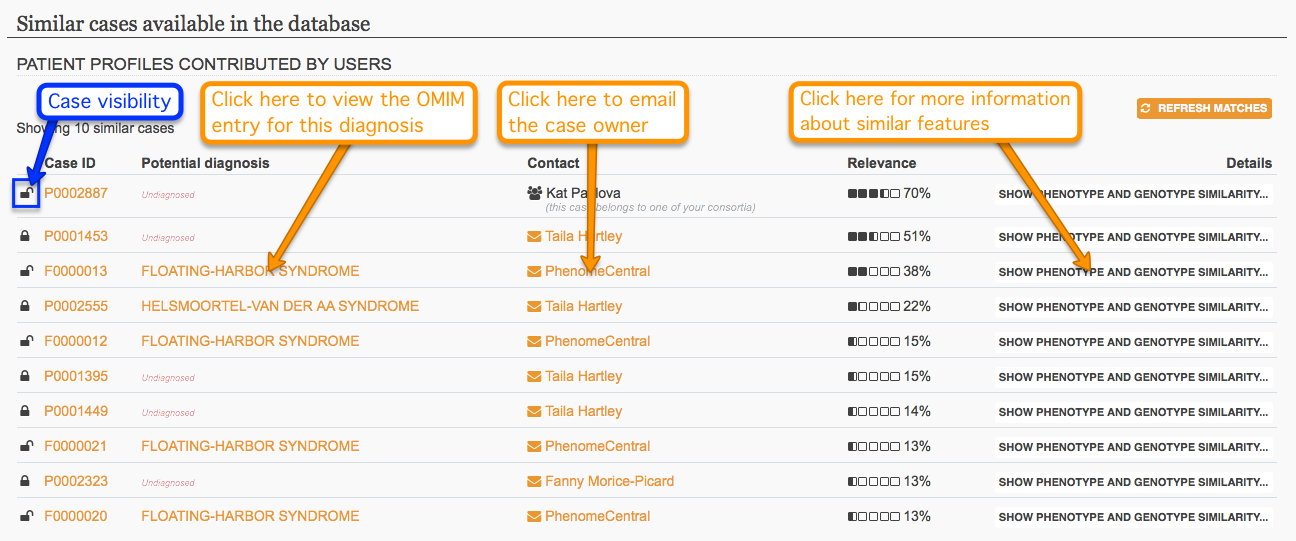
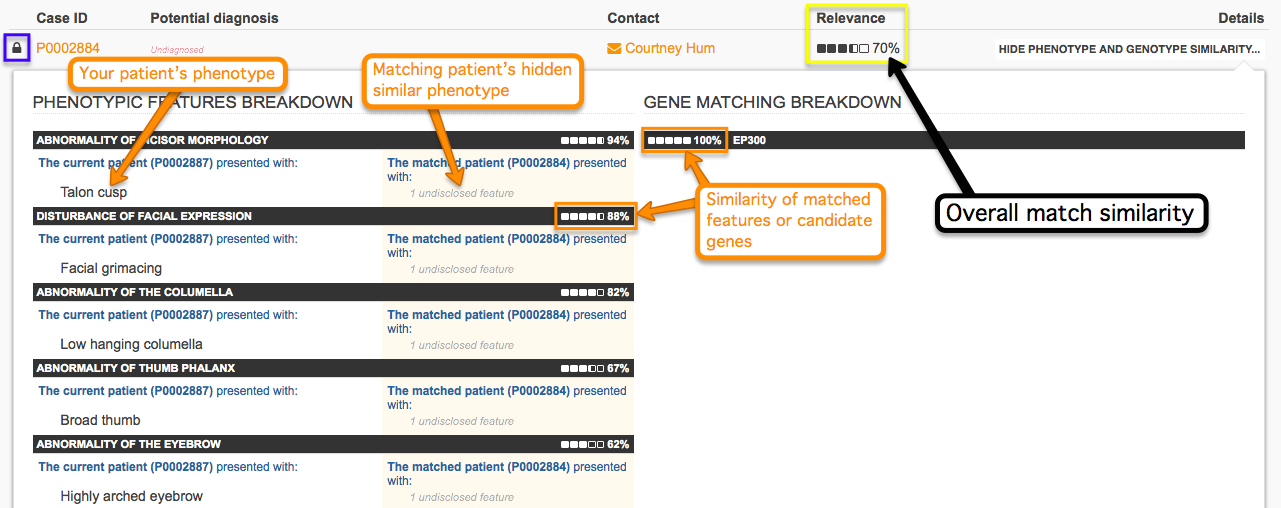
When comparing phenotype and genotype similarity between two cases, keep in mind that the information available for the matching case depends upon visibility level. Public cases that are similar are displayed without restrictions, while for matchable cases the features that have contributed to the high similarity score are hinted at without disclosing specific information.
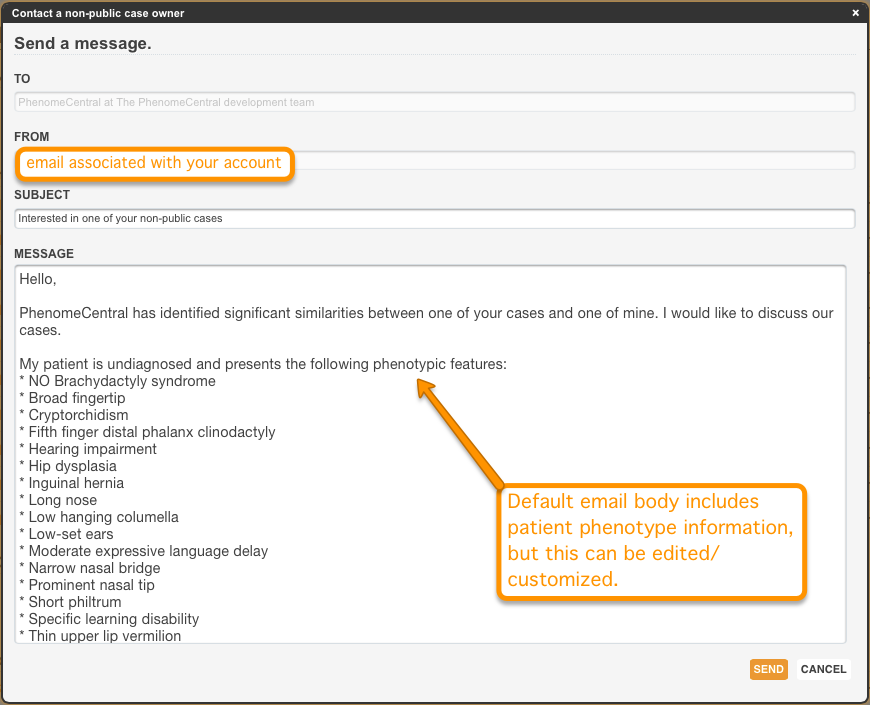
Establishing contact with owners of public and non-public similar cases
Selecting the case Contact opens a popup through which the case owner can be reached by email. The owner may choose to answer or ignore this discussion request. In this way, owners of cases set to matchable can become collaborators on cases of mutual interest, and can allow one another varying levels of access to individual cases.
Discovering similar cases in partner MME databases
PhenomeCentral is a founding member of the Matchmaker Exchange (MME), a collaborative effort to address the common challenge of exome and genome sequencing in both the research and clinical settings. As part of the MME, PhenomeCentral can query other member databases for similar patients. Currently, PhenomeCentral is connected to:
- DECIPHER
- GeneMatcher
- Australian Genomics Health Alliance Patient Archive
- Monarch Initiative (model organisms)
- Broad Institute matchbox
- MyGene2
- PubCaseFinder (disease literature)
It is not necessary for PhenomeCentral users to create accounts on these databases to discover matching patient profiles.
When first creating a patient record, you will be presented with 5 checkboxes for confirming patient consent. The 5th box must be selected in order to have your patient match with any patients over the MME. After creating a patient record and selecting Save and View Summary, the data is compared against all other patient records in PhenomeCentral only. If you scroll to the bottom of the summary page, you will see a section for Matches in partner MME databases. By selecting Find matches, you can choose to have the data compared against patients deposited in each of the databases connected to PhenomeCentral through the MME.
PhenomeCentral exists thanks to
The Human Phenotype Ontology

PhenomeCentral collects phenotype data using the Human Phenotype Ontology (HPO), a standardized vocabulary of phenotypic abnormalities encountered in human disease.
Thanks to the detailed terminology and semantic organization of the HPO, PhenomeCentral can match phenotypic profiles even when there are no phenotypes in common.
Find out more...The Exomiser
The Exomiser software package annotates, filters, and prioritizes variants in a patient's exome based on their phenotype (encoded using the HPO).
PhenomeCentral depends on the Exomiser to analyze exome sequence data and prioritize genes for genomic matching.
Find out more...The Monarch Initiative

The Monarch Initiative builds and maintains a curated knowledge base that links genotype-phenotype associations across a large number of species (including humans).
The infrastructure and algorithms developed by the Monarch Initiative provide the foundation for much of the PhenomeCentral core functionality, including phenotype matching, variant prioritization, diagnosis suggestions, and case specificity evaluation.
Find out more...PhenoTips™
PhenomeCentral was built on top of PhenoTips, an open source software for collecting and analyzing phenotypic and genotypic information of patients with genetic disorders.
PhenomeCentral leverages the PhenoTips user interface for capturing standardized data and analyzing it real-time, as well as its secure collaboration and data sharing capabilities.
Find out more...Citing PhenomeCentral
If PhenomeCentral enables a research publication, the authors must acknowledge PhenomeCentral using the following wording: "This study makes use of data shared through the PhenomeCentral repository. Funding for PhenomeCentral was provided by Genome Canada and Canadian Institute of Health Research (CIHR).
To cite PhenomeCentral in a research paper, please refer the PhenomeCentral presentation article:
Buske OJ*, Girdea M*, Dumitriu S, Gallinger B, Hartley T, Trang H, Misyura A, Friedman T, Beaulieu C, Bone WP, Links AE, Washington NL, Haendel MA, Robinson PN, Boerkoel CF, Adams D, Gahl WA, Boycott KM, Brudno M. 2015. PhenomeCentral: A Portal for Phenotypic and Genotypic Matchmaking of Patients with Rare Genetic Diseases. Human Mutation, 36: 931–940. doi:10.1002/humu.22851 [BibTeX]
@article{buske2015phenomecentral,
title={{PhenomeCentral}: a portal for phenotypic and genotypic matchmaking of patients with rare genetic diseases},
author={Buske, Orion J and Girdea, Marta and Dumitriu, Sergiu and Gallinger, Bailey and Hartley, Taila and Trang, Heather and Misyura, Andriy and Friedman, Tal and Beaulieu, Chandree and Bone, William P and others},
journal={Human Mutation},
volume={36},
number={10},
pages={931--940},
year={2015},
publisher={Wiley Online Library}
}
More questions?
Please address your questions or report issues encountered while using PhenomeCentral to support@phenomecentral.org.
